tidymodels
Dr. D’Agostino McGowan
tidymodels
- Go to the sta-363-s20 GitHub organization and search for
appex-04-tidymodels - Go to RStudio Pro
- rstudio.hpc.ar53.wfu.edu:8787
- pw: R2D2Star!
tidymodels

- tidymodels is an opinionated collection of R packages designed for modeling and statistical analysis.
- All packages share an underlying philosophy and a common grammar.
Step 1: Specify the model
- Pick the model
Step 1: Specify the model
- Pick the model
- Set the engine
Specify the model
linear_reg() %>% set_engine("lm")Specify the model
linear_reg() %>% set_engine("glmnet")Specify the model
linear_reg() %>% set_engine("spark")Specify the model
decision_tree() %>% set_engine("ranger")Specify the model
- All available models:
https://tidymodels.github.io/parsnip/articles/articles/Models.html
Specify Model
Write a pipe that creates a model that uses lm() to fit a linear regression using tidymodels. Save it as lm_spec and look at the object. What does it return?
Hint: you'll need https://tidymodels.github.io/parsnip/articles/articles/Models.html
02:00
lm_spec <- linear_reg() %>% # Pick linear regression set_engine(engine = "lm") # set enginelm_spec## Linear Regression Model Specification (regression)## ## Computational engine: lmFit the data
- You can train your model using the
fit()function
fit(lm_spec, mpg ~ horsepower, data = Auto)## parsnip model object## ## Fit time: 7ms ## ## Call:## stats::lm(formula = formula, data = data)## ## Coefficients:## (Intercept) horsepower ## 39.9359 -0.1578 Fit Model
Fit the model:
library(ISLR)lm_fit <- fit(lm_spec, mpg ~ horsepower, data = Auto)lm_fitDoes this give the same results as
lm(mpg ~ horsepower, data = Auto)01:30
Get predictions
lm_fit %>% predict(new_data = Auto)Get predictions
lm_fit %>% predict(new_data = Auto)- Still uses the
predict()function
Get predictions
lm_fit %>% predict(new_data = Auto)- Still uses the
predict()function - ‼️ Now
new_datahas an underscore
Get predictions
lm_fit %>% predict(new_data = Auto)- Still uses the
predict()function - ‼️ Now
new_datahas an underscore - 😄 This automagically creates a data frame
Get predictions
lm_fit %>% predict(new_data = Auto) %>% bind_cols(Auto)## # A tibble: 392 x 10## .pred mpg cylinders displacement horsepower weight acceleration year## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 19.4 18 8 307 130 3504 12 70## 2 13.9 15 8 350 165 3693 11.5 70## 3 16.3 18 8 318 150 3436 11 70## 4 16.3 16 8 304 150 3433 12 70## 5 17.8 17 8 302 140 3449 10.5 70## 6 8.68 15 8 429 198 4341 10 70## 7 5.21 14 8 454 220 4354 9 70## 8 6.00 14 8 440 215 4312 8.5 70## 9 4.42 14 8 455 225 4425 10 70## 10 9.95 15 8 390 190 3850 8.5 70## # … with 382 more rows, and 2 more variables: origin <dbl>, name <fct>01:30
Get predictions
Edit the code below to add the original data to the predicted data.
mpg_pred <- lm_fit %>% predict(new_data = Auto) %>% ---Get predictions
mpg_pred <- lm_fit %>% predict(new_data = Auto) %>% bind_cols(Auto)mpg_pred## # A tibble: 392 x 10## .pred mpg cylinders displacement horsepower weight acceleration year## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 19.4 18 8 307 130 3504 12 70## 2 13.9 15 8 350 165 3693 11.5 70## 3 16.3 18 8 318 150 3436 11 70## 4 16.3 16 8 304 150 3433 12 70## 5 17.8 17 8 302 140 3449 10.5 70## 6 8.68 15 8 429 198 4341 10 70## 7 5.21 14 8 454 220 4354 9 70## 8 6.00 14 8 440 215 4312 8.5 70## 9 4.42 14 8 455 225 4425 10 70## 10 9.95 15 8 390 190 3850 8.5 70## # … with 382 more rows, and 2 more variables: origin <dbl>, name <fct>Calculate the error
- Root mean square error
mpg_pred %>% rmse(truth = mpg, estimate = .pred)## # A tibble: 1 x 3## .metric .estimator .estimate## <chr> <chr> <dbl>## 1 rmse standard 4.89Calculate the error
- Root mean square error
mpg_pred %>% rmse(truth = mpg, estimate = .pred)## # A tibble: 1 x 3## .metric .estimator .estimate## <chr> <chr> <dbl>## 1 rmse standard 4.89What is this estimate? (training error? testing error?)
Validation set approach
Auto_split <- initial_split(Auto, prop = 0.5)Auto_split## <196/196/392>Validation set approach
Auto_split <- initial_split(Auto, prop = 0.5)Auto_split## <196/196/392>- Extract the training and testing data
training(Auto_split)testing(Auto_split)Validation set approach
Auto_train <- training(Auto_split)Auto_train## # A tibble: 196 x 9## mpg cylinders displacement horsepower weight acceleration year origin## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 18 8 307 130 3504 12 70 1## 2 17 8 302 140 3449 10.5 70 1## 3 15 8 429 198 4341 10 70 1## 4 14 8 454 220 4354 9 70 1## 5 14 8 440 215 4312 8.5 70 1## 6 14 8 455 225 4425 10 70 1## 7 15 8 390 190 3850 8.5 70 1## 8 14 8 340 160 3609 8 70 1## 9 14 8 455 225 3086 10 70 1## 10 24 4 113 95 2372 15 70 3## # … with 186 more rows, and 1 more variable: name <fct>04:00
Validation Set
Copy the code below, fill in the blanks to fit a model on the training data then calculate the test RMSE.
set.seed(100)Auto_split <- ________Auto_train <- ________Auto_test <- ________lm_fit <- fit(lm_spec, mpg ~ horsepower, data = ________)mpg_pred <- ________ %>% predict(new_data = ________) %>% bind_cols(________)rmse(________, truth = ________, estimate = ________)A faster way!
- You can use
last_fit()and specify the split - This will automatically train the data on the
traindata from the split - Instead of specifying which metric to calculate (with
rmseas before) you can just usecollect_metrics()and it will automatically calculate the metrics on thetestdata from the split
set.seed(100)Auto_split <- initial_split(Auto, prop = 0.5)lm_fit <- last_fit(lm_spec, mpg ~ horsepower, split = Auto_split)lm_fit %>% collect_metrics()## # A tibble: 2 x 3## .metric .estimator .estimate## <chr> <chr> <dbl>## 1 rmse standard 4.87 ## 2 rsq standard 0.625What about cross validation?
Auto_cv <- vfold_cv(Auto, v = 5)Auto_cv## # 5-fold cross-validation ## # A tibble: 5 x 2## splits id ## <named list> <chr>## 1 <split [313/79]> Fold1## 2 <split [313/79]> Fold2## 3 <split [314/78]> Fold3## 4 <split [314/78]> Fold4## 5 <split [314/78]> Fold5What about cross validation?
What about cross validation?
fit_resamples(lm_spec, mpg ~ horsepower, resamples = Auto_cv)What about cross validation?
fit_resamples(lm_spec, mpg ~ horsepower, resamples = Auto_cv)## # 5-fold cross-validation ## # A tibble: 5 x 4## splits id .metrics .notes ## * <list> <chr> <list> <list> ## 1 <split [313/79]> Fold1 <tibble [2 × 3]> <tibble [0 × 1]>## 2 <split [313/79]> Fold2 <tibble [2 × 3]> <tibble [0 × 1]>## 3 <split [314/78]> Fold3 <tibble [2 × 3]> <tibble [0 × 1]>## 4 <split [314/78]> Fold4 <tibble [2 × 3]> <tibble [0 × 1]>## 5 <split [314/78]> Fold5 <tibble [2 × 3]> <tibble [0 × 1]>What about cross validation?
How do we get the metrics out? With collect_metrics() again!
What about cross validation?
How do we get the metrics out? With collect_metrics() again!
results <- fit_resamples(lm_spec, mpg ~ horsepower, resamples = Auto_cv)results %>% collect_metrics()## # A tibble: 2 x 5## .metric .estimator mean n std_err## <chr> <chr> <dbl> <int> <dbl>## 1 rmse standard 4.93 5 0.0779## 2 rsq standard 0.611 5 0.027702:00
K-fold cross validation
Edit the code below to get the 5-fold cross validation error rate for the following model:
mpg=β0+β1horsepower+β2horsepower2+ϵ
Auto_cv <- vfold_cv(Auto, v = 5)results <- fit_resamples(lm_spec, ----, resamples = ---)results %>% collect_metrics()- What do you think
rsqis?
What if we wanted to do some preprocessing
- For the shrinkage methods we discussed it was important to scale the variables
What if we wanted to do some preprocessing
- For the shrinkage methods we discussed it was important to scale the variables
What does this mean?
What if we wanted to do some preprocessing
- For the shrinkage methods we discussed it was important to scale the variables
What does this mean?
What would happen if we scale before doing cross-validation? Will we get different answers?
What if we wanted to do some preprocessing
Auto_scaled <- Auto %>% mutate(horsepower = scale(horsepower))sd(Auto_scaled$horsepower)## [1] 1Auto_cv_scaled <- vfold_cv(Auto_scaled, v = 5)map_dbl(Auto_cv_scaled$splits, function(x) { dat <- as.data.frame(x)$horsepower sd(dat) })## 1 2 3 4 5 ## 0.9767551 0.9880625 1.0205261 0.9833461 1.0307327What if we wanted to do some preprocessing
recipe()!
What if we wanted to do some preprocessing
recipe()!- Using the
recipe()function along withstep_*()functions, we can specify preprocessing steps and R will automagically apply them to each fold appropriately.
What if we wanted to do some preprocessing
recipe()!- Using the
recipe()function along withstep_*()functions, we can specify preprocessing steps and R will automagically apply them to each fold appropriately.
rec <- recipe(mpg ~ horsepower, data = Auto) %>% step_scale(horsepower)What if we wanted to do some preprocessing
recipe()!- Using the
recipe()function along withstep_*()functions, we can specify preprocessing steps and R will automagically apply them to each fold appropriately.
rec <- recipe(mpg ~ horsepower, data = Auto) %>% step_scale(horsepower)- You can find all of the potential preprocessing steps here: https://tidymodels.github.io/recipes/reference/index.html
Where do we plug in this recipe?
- The
recipegets plugged into thefit_resamples()function
Where do we plug in this recipe?
- The
recipegets plugged into thefit_resamples()function
Auto_cv <- vfold_cv(Auto, v = 5)rec <- recipe(mpg ~ horsepower, data = Auto) %>% step_scale(horsepower)results <- fit_resamples(lm_spec, preprocessor = rec, resamples = Auto_cv)results %>% collect_metrics()## # A tibble: 2 x 5## .metric .estimator mean n std_err## <chr> <chr> <dbl> <int> <dbl>## 1 rmse standard 4.90 5 0.198 ## 2 rsq standard 0.608 5 0.0162What if we want to predict mpg with more variables
- Now we still want to add a step to scale predictors
- We could either write out all predictors individually to scale them
What if we want to predict mpg with more variables
- Now we still want to add a step to scale predictors
- We could either write out all predictors individually to scale them
- OR we could use the
all_predictors()short hand.
What if we want to predict mpg with more variables
- Now we still want to add a step to scale predictors
- We could either write out all predictors individually to scale them
- OR we could use the
all_predictors()short hand.
rec <- recipe(mpg ~ horsepower + displacement + weight, data = Auto) %>% step_scale(all_predictors())Putting it together
rec <- recipe(mpg ~ horsepower + displacement + weight, data = Auto) %>% step_scale(all_predictors())results <- fit_resamples(lm_spec, preprocessor = rec, resamples = Auto_cv)results %>% collect_metrics()## # A tibble: 2 x 5## .metric .estimator mean n std_err## <chr> <chr> <dbl> <int> <dbl>## 1 rmse standard 4.26 5 0.102 ## 2 rsq standard 0.711 5 0.0104Ridge, Lasso, and Elastic net
- When specifying your model, you can indicate whether you would like to use ridge, lasso, or elastic net. We can write a general equation to minimize:
RSS+λ((1−α)p∑i=1β2j+αp∑i=1|βj|)
Ridge, Lasso, and Elastic net
- When specifying your model, you can indicate whether you would like to use ridge, lasso, or elastic net. We can write a general equation to minimize:
RSS+λ((1−α)p∑i=1β2j+αp∑i=1|βj|)
lm_spec <- linear_reg() %>% set_engine("glmnet")- First specify the engine. We'll use
glmnet
Ridge, Lasso, and Elastic net
- When specifying your model, you can indicate whether you would like to use ridge, lasso, or elastic net. We can write a general equation to minimize:
RSS+λ((1−α)p∑i=1β2j+αp∑i=1|βj|)
lm_spec <- linear_reg() %>% set_engine("glmnet")- First specify the engine. We'll use
glmnet - The
linear_reg()function has two additional parameters,penaltyandmixture
Ridge, Lasso, and Elastic net
- When specifying your model, you can indicate whether you would like to use ridge, lasso, or elastic net. We can write a general equation to minimize:
RSS+λ((1−α)p∑i=1β2j+αp∑i=1|βj|)
lm_spec <- linear_reg() %>% set_engine("glmnet")- First specify the engine. We'll use
glmnet - The
linear_reg()function has two additional parameters,penaltyandmixture penaltyis λ from our equation.
Ridge, Lasso, and Elastic net
- When specifying your model, you can indicate whether you would like to use ridge, lasso, or elastic net. We can write a general equation to minimize:
RSS+λ((1−α)p∑i=1β2j+αp∑i=1|βj|)
lm_spec <- linear_reg() %>% set_engine("glmnet")- First specify the engine. We'll use
glmnet - The
linear_reg()function has two additional parameters,penaltyandmixture penaltyis λ from our equation.mixtureis a number between 0 and 1 representing α
Ridge, Lasso, and Elastic net
RSS+λ((1−α)p∑i=1β2j+αp∑i=1|βj|)
What would we set mixture to in order to perform Ridge regression?
Ridge, Lasso, and Elastic net
RSS+λ((1−α)p∑i=1β2j+αp∑i=1|βj|)
What would we set mixture to in order to perform Ridge regression?
ridge_spec <- linear_reg(penalty = 100, mixture = 0) %>% set_engine("glmnet")02:00
Lasso specification
Set up the model specification to fit a Lasso with a λ value of 5. Call this object lasso_spec.
Ridge, Lasso, and Elastic net
RSS+λ((1−α)p∑i=1β2j+αp∑i=1|βj|)
ridge_spec <- linear_reg(penalty = 100, mixture = 0) %>% set_engine("glmnet")Ridge, Lasso, and Elastic net
RSS+λ((1−α)p∑i=1β2j+αp∑i=1|βj|)
ridge_spec <- linear_reg(penalty = 100, mixture = 0) %>% set_engine("glmnet")lasso_spec <- linear_reg(penalty = 5, mixture = 1) %>% set_engine("glmnet")Ridge, Lasso, and Elastic net
RSS+λ((1−α)p∑i=1β2j+αp∑i=1|βj|)
ridge_spec <- linear_reg(penalty = 100, mixture = 0) %>% set_engine("glmnet")lasso_spec <- linear_reg(penalty = 5, mixture = 1) %>% set_engine("glmnet")enet_spec <- linear_reg(penalty = 60, mixture = 0.7) %>% set_engine("glmnet")Okay, but we wanted to look at 3 different models!
ridge_spec <- linear_reg(penalty = 100, mixture = 0) %>% set_engine("glmnet") results <- fit_resamples(ridge_spec, preprocessor = rec, resamples = Auto_cv)Okay, but we wanted to look at 3 different models!
ridge_spec <- linear_reg(penalty = 100, mixture = 0) %>% set_engine("glmnet") results <- fit_resamples(ridge_spec, preprocessor = rec, resamples = Auto_cv)lasso_spec <- linear_reg(penalty = 5, mixture = 1) %>% set_engine("glmnet") results <- fit_resamples(lasso_spec, preprocessor = rec, resamples = Auto_cv)elastic_spec <- linear_reg(penalty = 60, mixture = 0.7) %>% set_engine("glmnet") results <- fit_resamples(elastic_spec, preprocessor = rec, resamples = Auto_cv)elastic_spec <- linear_reg(penalty = 60, mixture = 0.7) %>% set_engine("glmnet") results <- fit_resamples(elastic_spec, preprocessor = rec, resamples = Auto_cv)- 😱 this looks like copy + pasting!
tune 🎶
penalty_spec <- linear_reg(penalty = tune(), mixture = tune()) %>% set_engine("glmnet")- Notice the code above has
tune()for the the penalty and the mixture. Those are the things we want to vary!
tune 🎶
- Now we need to create a grid of potential penalties ( λ ) and mixtures ( α ) that we want to test
- Instead of
fit_resamples()we are going to usetune_grid()
grid <- expand_grid(penalty = seq(0, 100, by = 10), mixture = seq(0, 1, by = 0.2))results <- tune_grid(penalty_spec, preprocessor = rec, grid = grid, resamples = Auto_cv)tune 🎶
results %>% collect_metrics()## # A tibble: 132 x 7## penalty mixture .metric .estimator mean n std_err## <dbl> <dbl> <chr> <chr> <dbl> <int> <dbl>## 1 0 0 rmse standard 4.28 5 0.118 ## 2 0 0 rsq standard 0.709 5 0.00929## 3 0 0.2 rmse standard 4.26 5 0.103 ## 4 0 0.2 rsq standard 0.711 5 0.0102 ## 5 0 0.4 rmse standard 4.26 5 0.104 ## 6 0 0.4 rsq standard 0.711 5 0.0103 ## 7 0 0.6 rmse standard 4.26 5 0.105 ## 8 0 0.6 rsq standard 0.711 5 0.0103 ## 9 0 0.8 rmse standard 4.26 5 0.106 ## 10 0 0.8 rsq standard 0.711 5 0.0103 ## # … with 122 more rowsSubset results
results %>% collect_metrics() %>% filter(.metric == "rmse") %>% arrange(mean)## # A tibble: 66 x 7## penalty mixture .metric .estimator mean n std_err## <dbl> <dbl> <chr> <chr> <dbl> <int> <dbl>## 1 0 0.6 rmse standard 4.26 5 0.105## 2 0 0.8 rmse standard 4.26 5 0.106## 3 0 0.2 rmse standard 4.26 5 0.103## 4 0 0.4 rmse standard 4.26 5 0.104## 5 0 1 rmse standard 4.26 5 0.106## 6 0 0 rmse standard 4.28 5 0.118## 7 10 0 rmse standard 4.76 5 0.244## 8 20 0 rmse standard 5.31 5 0.265## 9 10 0.2 rmse standard 5.39 5 0.266## 10 30 0 rmse standard 5.72 5 0.262## # … with 56 more rows- Since this is a data frame, we can do things like filter and arrange!
Subset results
results %>% collect_metrics() %>% filter(.metric == "rmse") %>% arrange(mean)## # A tibble: 66 x 7## penalty mixture .metric .estimator mean n std_err## <dbl> <dbl> <chr> <chr> <dbl> <int> <dbl>## 1 0 0.6 rmse standard 4.26 5 0.105## 2 0 0.8 rmse standard 4.26 5 0.106## 3 0 0.2 rmse standard 4.26 5 0.103## 4 0 0.4 rmse standard 4.26 5 0.104## 5 0 1 rmse standard 4.26 5 0.106## 6 0 0 rmse standard 4.28 5 0.118## 7 10 0 rmse standard 4.76 5 0.244## 8 20 0 rmse standard 5.31 5 0.265## 9 10 0.2 rmse standard 5.39 5 0.266## 10 30 0 rmse standard 5.72 5 0.262## # … with 56 more rows- Since this is a data frame, we can do things like filter and arrange!
Which would you choose?
results %>% collect_metrics() %>% filter(.metric == "rmse") %>% ggplot(aes(penalty, mean, color = factor(mixture), group = factor(mixture))) + geom_line() + geom_point() + labs(y = "RMSE")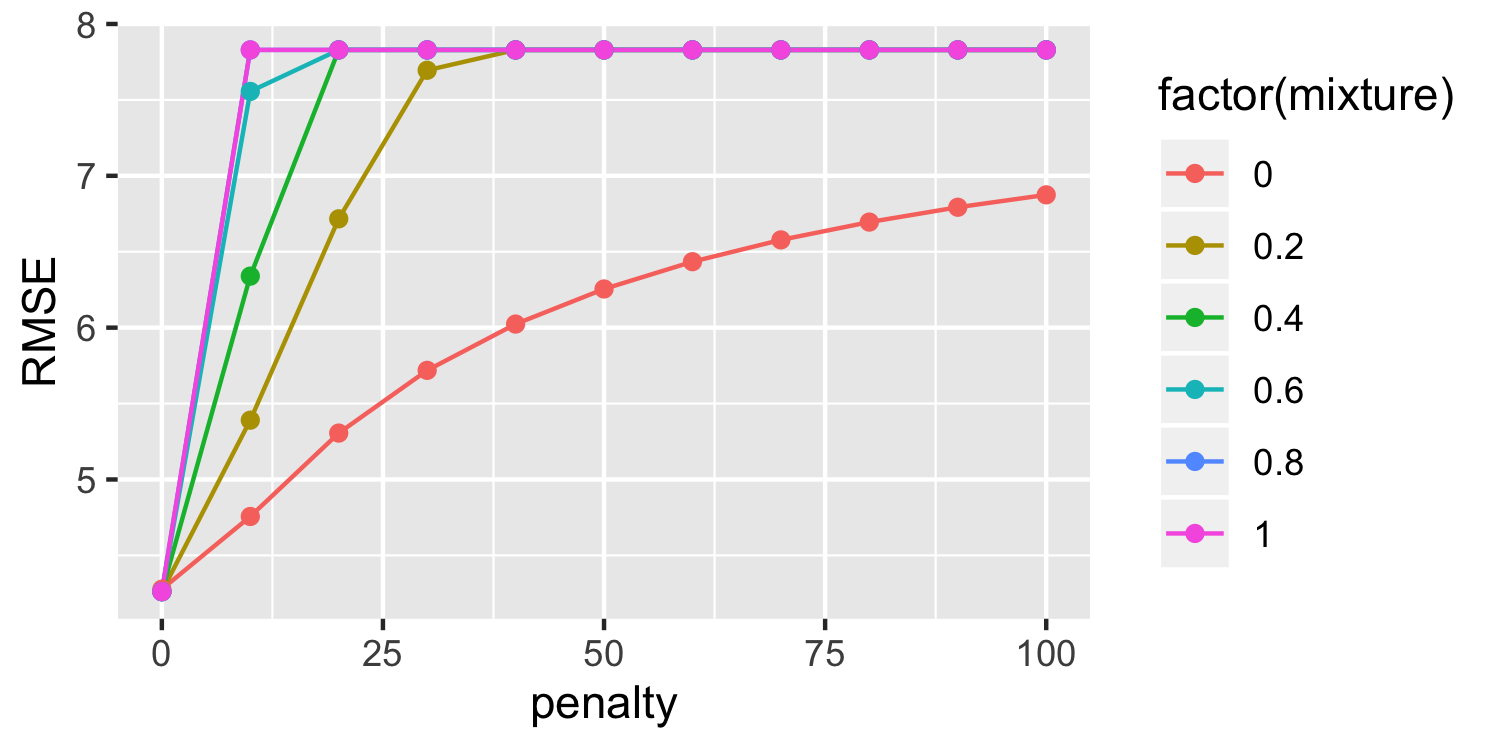
Putting it all together
- Often we can use a combination of all of these tools together
- First split our data
- Do cross validation on just the training data to tune the parameters
- Use
last_fit()with the selected parameters, specifying the split data so that it is evaluated on the left out test sample
Putting it all together
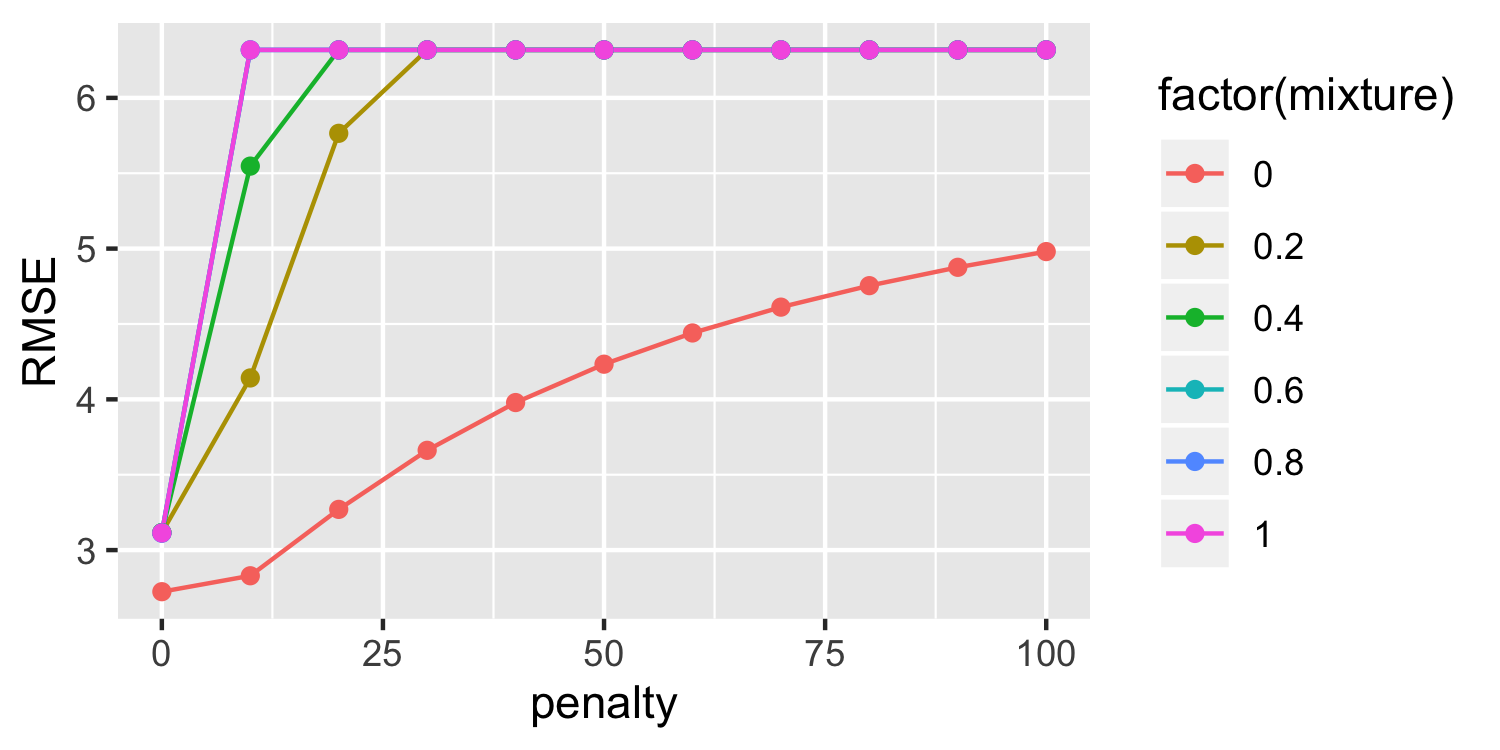
auto_split <- initial_split(Auto, prop = 0.5)auto_train <- training(auto_split)auto_cv <- vfold_cv(auto_train, v = 5)rec <- recipe(mpg ~ horsepower + displacement + weight, data = auto_train) %>% step_scale(all_predictors())tuning <- tune_grid(penalty_spec, rec, grid = grid, resamples = auto_cv)tuning %>% collect_metrics() %>% filter(.metric == "rmse") %>% arrange(mean)## # A tibble: 66 x 7## penalty mixture .metric .estimator mean n std_err## <dbl> <dbl> <chr> <chr> <dbl> <int> <dbl>## 1 0 1 rmse standard 4.15 5 0.267## 2 0 0.8 rmse standard 4.15 5 0.267## 3 0 0.6 rmse standard 4.15 5 0.266## 4 0 0.4 rmse standard 4.15 5 0.266## 5 0 0.2 rmse standard 4.15 5 0.266## 6 0 0 rmse standard 4.17 5 0.276## 7 10 0 rmse standard 4.60 5 0.428## 8 20 0 rmse standard 5.11 5 0.476## 9 10 0.2 rmse standard 5.22 5 0.490## 10 30 0 rmse standard 5.50 5 0.490## # … with 56 more rowsPutting it all together
final_spec <- linear_reg(penalty = 0, mixture = 0) %>% set_engine("glmnet")fit <- last_fit(final_spec, rec, split = auto_split)fit %>% collect_metrics()## # A tibble: 2 x 3## .metric .estimator .estimate## <chr> <chr> <dbl>## 1 rmse standard 4.43 ## 2 rsq standard 0.711